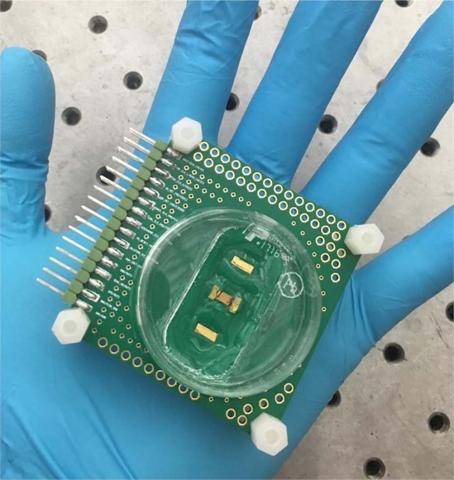
An electronic sensor platform that measures multi-physical cellular responses could reduce costs and cut time for new drug development.
Finding ways to improve the drug development process – which is currently costly, time-consuming and has an astronomically high failure rate – could have far-reaching benefits for health care and the economy. Researchers from the Georgia Institute of Technology have designed a cellular interfacing array using low-cost electronics that measures multiple cellular properties and responses in real time. This could enable many more potential drugs to be comprehensively tested for efficacy and toxic effects much faster. That’s why Hua Wang, associate professor in the School of Electrical and Computer Engineering at Georgia Tech, describes it as “helping us find the golden needle in the haystack.”
Pharmaceutical companies use cell-based assays, a combination of living cells and sensor electronics, to measure physiological changes in the cells. That data is used for high-throughput screening (HTS) during drug discovery. In this early phase of drug development, the goal is to identify target pathways and promising chemical compounds that could be developed further – and to eliminate those that are ineffective or toxic – by measuring the physiological responses of the cells to each compound.
Phenotypic testing of thousands of candidate compounds, with the majority “failing early,” allows only the most promising ones to be further developed into drugs and maybe eventually to undergo clinical trials, where drug failure is much more costly. But most existing cell-based assays use electronic sensors that can only measure one physiological property at a time and cannot obtain holistic cellular responses.
That’s where the new cellular sensing platform comes in. “The innovation of our technology is that we are able to leverage the advance of nano-electronic technologies to create cellular interfacing platforms with massively parallel pixels,” said Wang. “And within each pixel we can detect multiple physiological parameters from the same group of cells at the same time.” The experimental quad-modality chip features extracellular or intracellular potential recording, optical detection, cellular impedance measurement, and biphasic current stimulation.
Wang said the new technology offers four advantages over existing platforms:
Multimodal sensing: The chip’s ability to record multiple parameters on the same cellular sample gives researchers the ability to comprehensively monitor complex cellular responses, uncover the correlations among those parameters and investigate how they may respond together when exposed to drugs. “Living cells are small but highly complex systems. Drug administration often results in multiple physiological changes, but this cannot be detected using conventional single-modal sensing,” said Wang.
Large field of view: The platform allows researchers to examine the behavior of cells in a large aggregate to see how they respond collectively at the tissue level.
Small spatial resolution: Not only can researchers look at cells at the tissue level, they could also examine them at single-cell or even sub-cellular resolution.
Low-cost platform: The new array platform is built on standard complementary metal oxide semiconductor (CMOS) technologies, which is also used to build computer chips, and can be easily scaled up for mass production.
Wang’s team worked closely with Hee Cheol Cho, associate professor and the Urowsky-Sahr Scholar in Pediatric Bioengineering, whose Heart Regeneration lab is part of the Wallace Coulter Department of Biomedical Engineering at Georgia Tech and Emory University. They used neonatal rat ventricular myocytes and cardiac fibroblasts to illustrate the multi-parametric cell profiling ability of the array for drug screening. The recent results were published in the Royal Society of Chemistry’s journal Lab on a Chip on August 31, 2018.
Monitoring cellular responses in multi-physical domains and holistic multi-parametric cellular profiling should also prove beneficial in screening out chemical compounds that could have harmful effects on certain organs, said Jong Seok Park, a post-doctoral fellow in Wang’s lab and a leading author of the study. Many drugs have been withdrawn from the market after discoveries that they had toxic effects on the heart or liver, for example. This platform should enable researchers to comprehensively test for organ toxicity and other side effects at the initial phases of drug discovery.
The experimental chip may be useful for other applications, including personalized medicine – for example, testing cancer cells from a particular patient. “Patient to patient variation is huge, even with the same type of drug,” said Wang. The cellular interface array could be used to see which combination of existing drugs would give the best response and to find the optimum dose that is most effective with minimum toxicity to healthy cells.
The chip is capable of actuation as well as sensing. In the future, Wang said that cellular data from the chip could be uploaded and processed, and based on that, commands for new actuation or data acquisition could be sent to the chip automatically and wirelessly. He envisions rooms and rooms containing culture chambers with millions of such chips in fully automated facilities, “just automatically doing new drug selection for us,” he said.
Beyond these applications, Wang noted the scientific value of the research itself. Integrated circuits and nanoelectronics are some of the most sophisticated technology platforms created by humans. Living cells, on the other hand, are complex products produced through billions of years of natural selection and evolution.
“The central theme of our research is how we can leverage the best platform created by nature with the best platform created by humans,” he said. “Can we let them work together to create hybrid systems that achieve capabilities beyond biology only or electronics only systems? The fundamental scientific question we are addressing is how we can let inorganic electronics better interface with organic living cells.”
These researchers also participated in the related studies: Doohwan Jung, Adam Wang, Taiyun Chi, Sensen Li and Moez K. Aziz from the School of Electrical and Computer Engineering at Georgia Tech; and Sandra I. Grijalva and Michael N. Sayegh from the Department of Biomedical Engineering at Emory University. The research was funded in part by the National Science Foundation CAREER Award and ECCS CCSS Program, National Science Foundation Graduate Research Fellowship grant numbers DGE-1148903 and DGE-1650044, Office of Naval Research, and Semiconductor Research Corporation SSB roadmap consortium. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the funding agencies.
CITATION: Jong Seok Park, et al., “Multi-parametric cell profiling with a CMOS quad-modality cellular interfacing array for label-free fully automated drug screening,” (Lab Chip 2018). DOI: 10.1039/c8lc00156a
Research News
Georgia Institute of Technology
177 North Avenue
Atlanta, Georgia 30332-0181 USA
Media Relations Contact: John Toon (404-894-6986) (john.toon@comm.gatech.edu).
Writer: Kenna Simmons
Additional Images