Ph.D. students Hansol Yoon and Kyungduck Yoon's method has been featured in Nature Communications for achieving real-time panoramic super-resolution, opening doors to advanced large-scale cellular analysis.
(text and background only visible when logged in)
(text and background only visible when logged in)
A new imaging technique developed at Georgia Tech, called super-resolution panoramic integration (SPI), is redefining what’s possible in biological microscopy.
The research, conducted by Hansol Yoon, a Ph.D. student in the School of Electrical and Computer Engineering and Kyungduck Yoon, a Ph.D. student in the George W. Woodruff School of Mechanical Engineering, who conducts research in the Parker H. Petit Institute for Bioengineering and Biosciences, was recently published in Nature Communications.
Working in the Laboratory for Systems Biophotonics led by Wallace H. Coulter Department of Biomedical Engineering Associate Professor Shu Jia, and collaborating with School of Biological Sciences Professor William Ratcliff, the team designed SPI to overcome long-standing trade-offs between resolution and speed that have limited traditional microscopy methods.
“It’s similar to the panorama mode on a smartphone camera,” Yoon said. “SPI continuously scans biological samples while generating panoramic, super-resolved images in real time.”
The innovation addresses a critical challenge in microscopy. While microscopes have long been a cornerstone of scientific discovery, many biological features remain beyond the reach of conventional optical system because they are smaller than the diffraction limit of light (about 200 nanometers).

(From left to right) Kyungduck Yoon and Hansol Yoon
Although the advent of super-resolution microscopy—recognized with the 2014 Nobel Prize in Chemistry—overcame this limitation, such techniques typically require long imaging times and are often unsuitable for large-scale cellular analyses.SPI changes that by combining optical processing at the speed of light for instantaneous super-resolution image formation with high-content sweeping and synchronized line-scan readout for panoramic imaging.
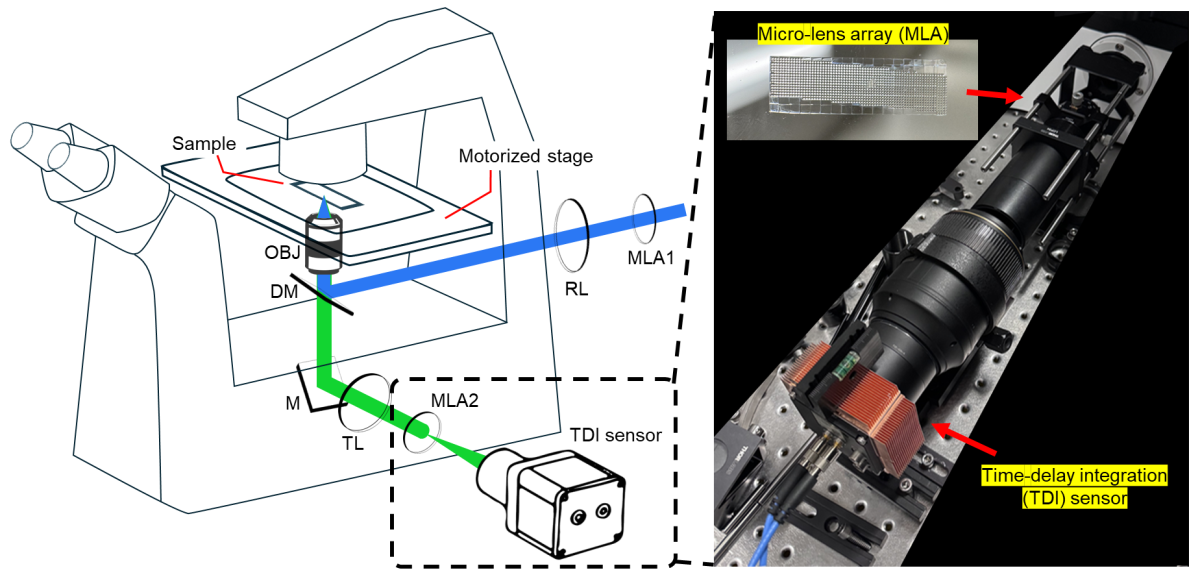
The SPI setup integrates easily into commonly used standard fluorescence microscopes.
The method integrates optical super-resolution processing with panoramic line-scan readout: a pair of microlens arrays optically processes fluorescence signals at the speed of light to form super-resolved images, while a time-delay integration (TDI) sensor synchronized with stage scanning integrates these images into seamless panoramic images.
Unlike conventional methods that provide either a broad overview or fine details, SPI enables large-scale, super-resolution cellular analysis with minimal post-processing and full compatibility with standard fluorescence microscopes commonly used in biology laboratories.
Demonstrated across diverse biological applications, SPI reveals subcellular and population-level morphology, function, and heterogeneity with unprecedented detail. This opens new opportunities for biological discovery beyond traditional optical and computational constraints, according to Yoon.
This method effectively scales super-resolution imaging to accommodate large cell populations and whole-slide applications, making it ideal for studies such as peripheral blood smear analysis. It also holds promise for evolutionary cell biology, enabling large-scale analyses while preserving subcellular precision.

SPI enables subcellular and cluster-level analysis of snowflake yeast populations at different evolutionary stages, from the ancestor to 1,000 days of evolution.
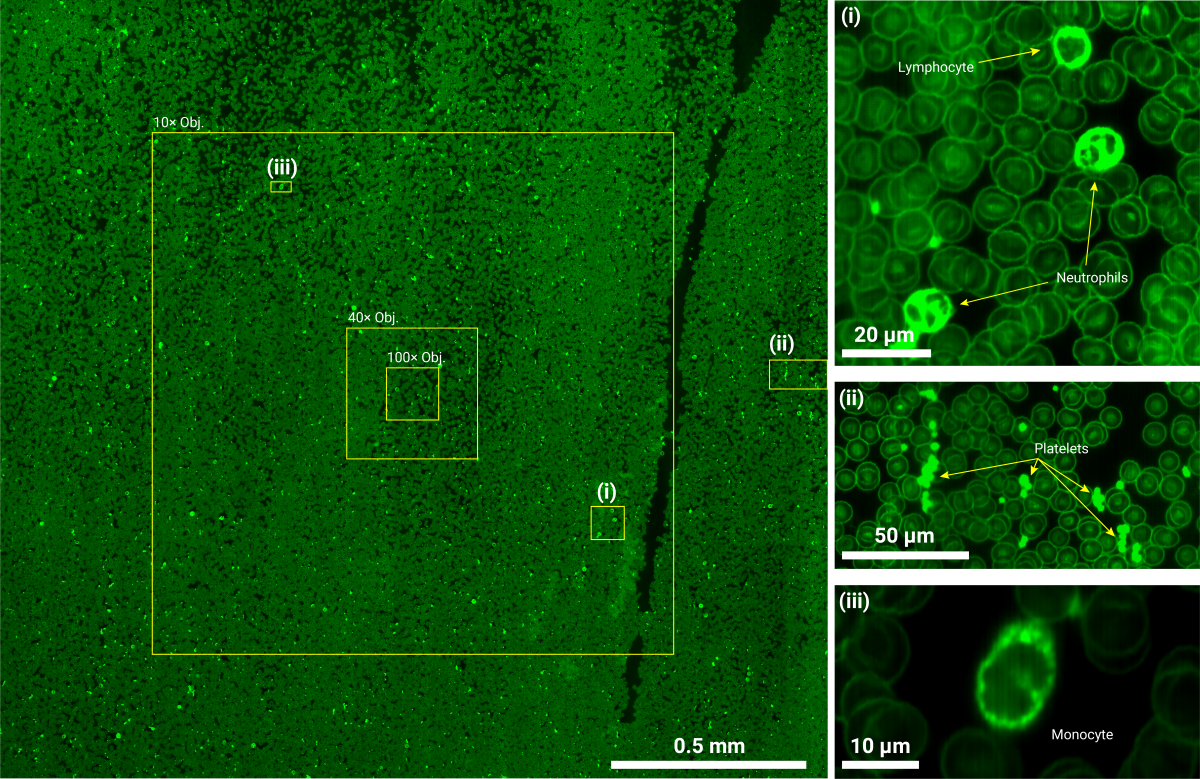
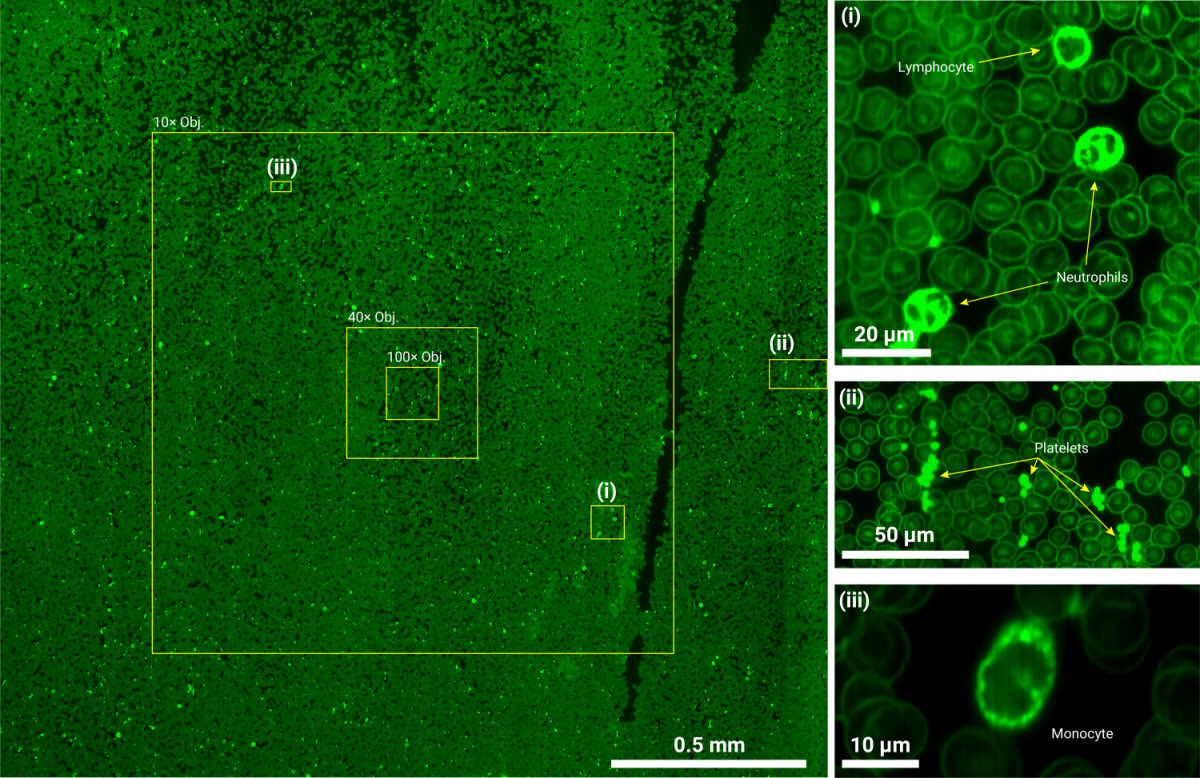
Super-resolution image of millimeter-scale blood smear regions, compared with static fields of view (10x, 40x, 100x) from conventional fluorescence microscopy.
With the promising results, the team is now working to secure a grant to advance SPI from a laboratory method to a commercial product.
Funding
This work is supported by the Parker H. Petit Institute for Bioengineering and Biosciences of Georgia Institute of Technology, the National Science Foundation grants 2503686 (to S.J.), 2145235 (to S.J.), and DEB-2452109 (to G.O.B.), the National Institutes of Health grants R35GM124846 (to S.J.) and R35GM138030 (to W.C.R.), and the Georgia Research Alliance grant GRA.26.005.GT.01.a. (to S.J.). This work was performed in part at the Georgia Tech Institute for Electronics and Nanotechnology, a member of the NSF-supported National Nanotechnology Coordinated Infrastructure (ECCS-1542174). We thank Dr. Parvin Forghani Esfahani of Dr. Chunhui Xu’s laboratory at Emory University for kindly providing human-induced pluripotent stem cell (hiPSC)-derived cardiac fibroblast samples used in this study. We also thank Nikolas Roeske at the Georgia Tech Institute for Electronics and Nanotechnology for his instruction and training in Nanoscribe 3D lithography, which enabled the fabrication of the custom microlens arrays.
Competing Interests
Shu Jia, Kyungduck Yoon., and Hansol Yoon are listed as inventors on a pending patent application (US Patent App. 63/814,286), partly relevant to the technology described in this manuscript.
Related Content
Tangolar Wins Best Paper Award for Non-Invasive Cardiac Output Monitoring
The research from Regents Entrepreneur Omer Inan’s research lab was recognized at the 2025 IEEE-EMBS International Conference on Body Sensor Networks for demonstrating how cardiac output can be measured with non-invasive cardiovascular signals.
Georgia Tech Brings Industry and Academia Together to Plot the Future of AI in Healthcare
The Accessible Healthcare through AI-Augmented Decisions (AHeAD) Center’s planning meeting charted a path to enhance patient experiences and outcomes through AI innovation.

